KIMMDY

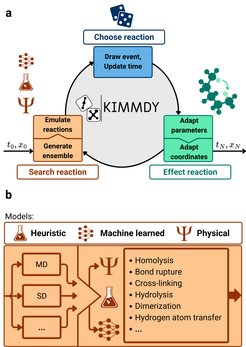
We have developed KIMMDY (Kinetic Monte Carlo / Molecular Dynamics), a hybrid simulation scheme that incorporates reactivity into Molecular Dynamics (MD) simulations using GROMACS. By combining MD with reactive Kinetic Monte Carlo (KMC) steps, KIMMDY enables the study of reaction dynamics, efficiently bridging time scales and overcoming conventional MD limitations.
Reactions are sampled based on their kinetic rates, which are computed using specialized reaction-specific plugins. Currently, KIMMDY supports rate predictions for homolytic bond rupture, hydrolysis, hydrogen atom transfers in collagen and alcohols and DNA dimerisation with additional reaction types and biomolecules under development. KIMMDY also takes full advantage of our machine learned Molecular Mechanics Force Field GRAPPA to predict simulation parameters for the dynamically changing topology.
KIMMDY is designed as a modular and extensible framework, allowing users to integrate custom plugins for additional reaction types and molecular systems.
Resources
You can find our code, documentation and handy tutorials on GitHub.
Read the detailed description of the method here.

