SAPs4Tissue: Self-assembling bioactive peptides for the biomimetic design of functional cell niches in human tissue models
In this 4-year cooperation program, jointly funded by the Fraunhofer and Max-Planck Societies, institutes from both societies push fundamental science towards real-life applications. Our group has teamed up with the Fraunhofer Institute for Silicate Research (Fh-ISC) and the Center for Translational Regenerative Therapies (TLZ-RT), Würzburg, to develop next generation biomaterials for human tissue models.
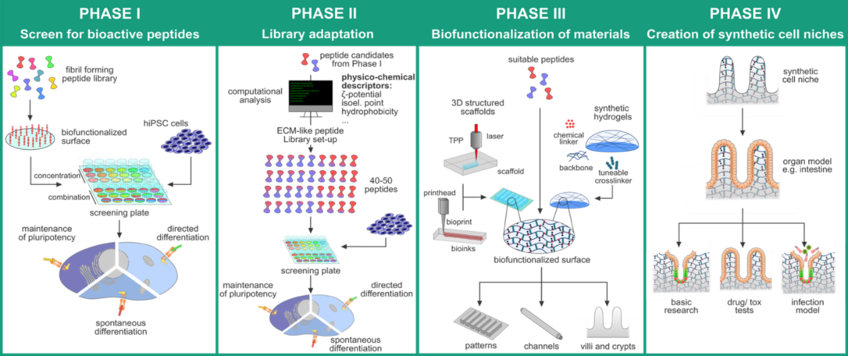
We will integrate essential characteristics of the native extracellular matrix (ECM) into biomaterial scaffolds to generate complex human tissue models, by enabling the material to communicate with the cell through relevant signals. Following the “function-by-design” principle, we will by synthesize, characterize, and screen a library of bioactive self-assembling peptides (SAPs) and generate bioactive peptide (co-)assemblies for the support of human cells. The combination of physico-chemical characterization and biological performance we obtain from the library will allow us to establish sequence-structure-bioactivity correlations. Computational methods, especially machine learning approaches will support the identification of relevant physico-chemical descriptors and may enable the prediction of highly bioactive peptide sequences. Next, bioactive peptides will be combined with both existing and novel biomaterial scaffolds to accurately reproduce the native cell niche. This requires additive manufacturing techniques, which are available through the Fh-ISC and TLZ-RT.
This project will create a fundamental insight into cell-material communication, structure-function relationships, and ultimately generate new, bioactive scaffold materials that will provide ideal cell niches for human tissue models.